Two stories describing two new tech developed in the lab are online at Nature Communications.
First paper (10.1038/s41467-019-12339-7), published in September, describes Casilio-ME – methylation editing modules for editing DNA methylation. To enhance DNA demethylation, we also used Casilio-sgRNA scaffold to assemble multiple enzymes at target loci to ‘streamline’ the multi-step DNA demethylation processes, resulting in enhanced DNA methylation. Congrats to Aziz Taghbalout who led the study; Menghan Du, UConn Health graduate student, Nate Jillette, our lab manager + RA, as well as our collaborators from the Sheng Li lab and Chris Heinen lab. A nice press release by Mark Wanner: https://bit.ly/36zM6fD
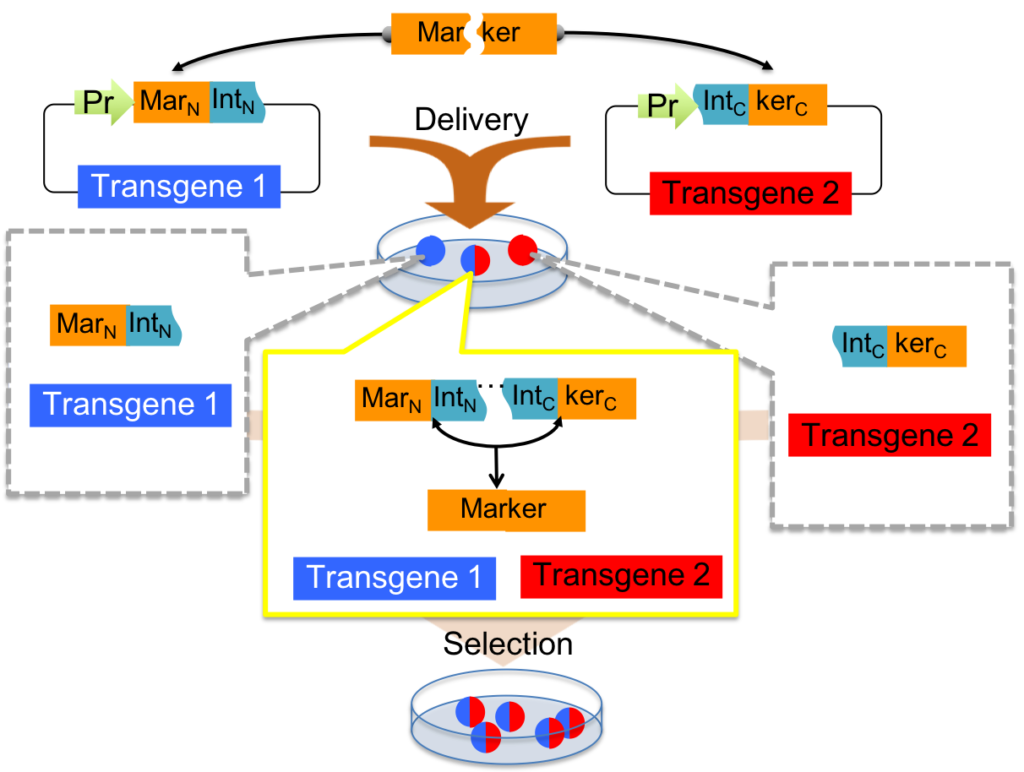
Second paper (10.1038/s41467-019-12891-2), published on the last day of October, describes split selectable markers wherein selectable markers such as antibiotic resistance genes, or fluorescent proteins, are divided into fragments fused to protein splicing elements called “inteins”, and are reconstituted in cells receiving all fragments during transgenesis. These allow co-selection of multiple transgenes using a selection scheme, increasing the capacity of transgenic selection. Congrats to co-first authors Nate Jillette (Lab Manager + RA) and Menghan Du (UConn Grad Student)! Also congrats to the team of Barbara Di Ventura from University of Freiburg for publishing back-to-back a similar story (10.1038/s41467-019-12911-1) describing similar strategies to split selectable markers. It feels so good to have a similar story published at the same time from another group! Press release by Mark Wanner: https://bit.ly/32nY7S7