Split Selectable Markers
Plasmids available on Genomics-online.com
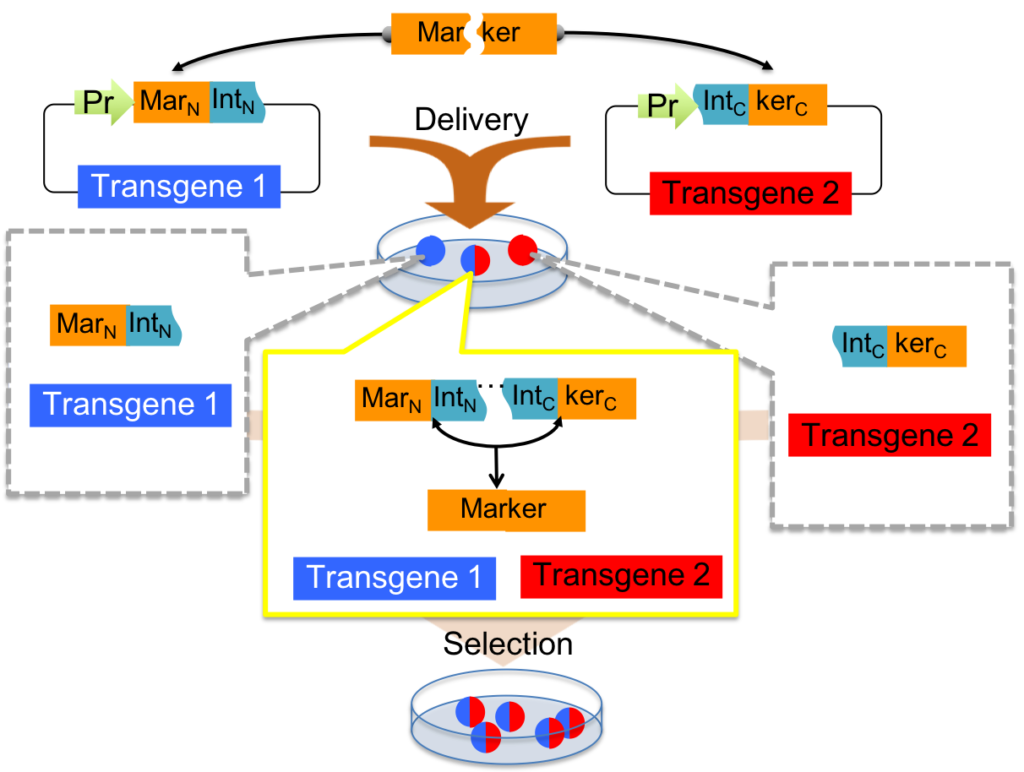
Split selectable markers. Selectable markers are widely used in transgenesis and genome editing for selecting engineered cells with a desired genotype but the variety of markers is limited. Split selectable markers are created by dividing fluorescent proteins or antibiotic resistance proteins into several segments fused to protein splicing elements called “inteins” can be separately co-segregated with different transgenic vectors, and rejoin via protein trans-splicing to reconstitute a full-length marker protein in host cells receiving all intended vectors. In a work led by lab manager/RA Nathaniel Jillette and PhD candidate graduate student Menghan Du, we demonstrated utilities of split selectable markers in lentiviral transgenesis and CRISPR/Cas-mediated genome editing.
▲Selection achieved by intein-split resistance (intres) genes.
▲Nate (left), Albert (center), Menghan (right). Photography by Charles Camarda.
Publication:
Jillette, N., Du, M., Zhu, J.J., Cardoz, P., Cheng, A.W. (2019) Split Selectable Markers. Nature Communications 10:4968. doi: 10.1038/s41467-019-12891-2 PMID: 31672965 PMCID: PMC6823381
Press release:
“Improving transgene research with split selectable markers” by Mark Wanner
Grant support:
R01HG009900, P30CA034196.